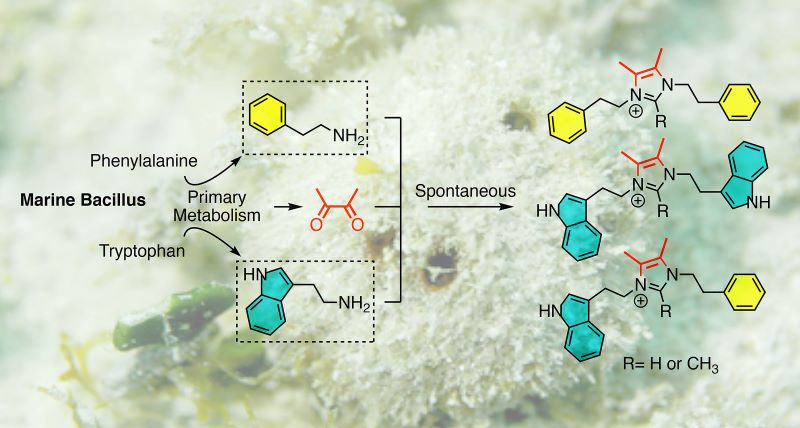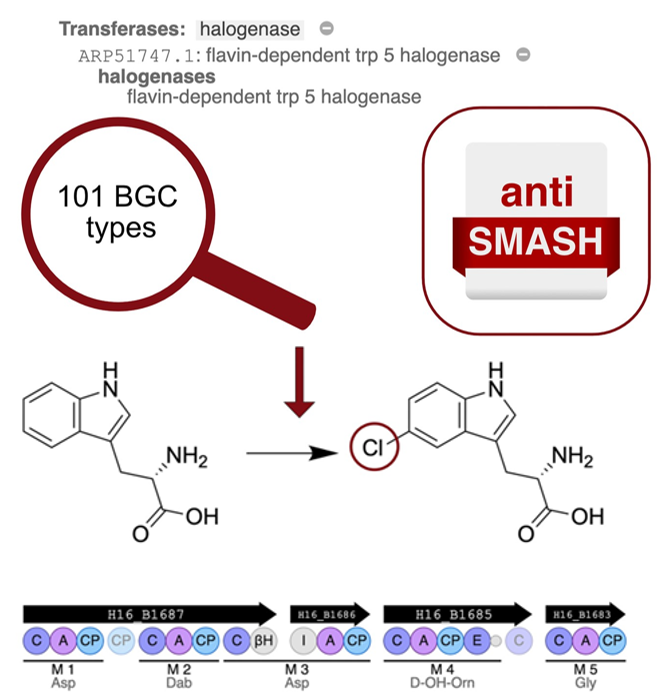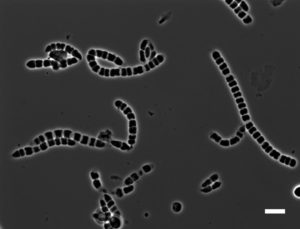
Insights into the lifestyle of uncultured bacterial natural product factories associated with marine sponges | Proceedings of the National Academy of Sciences
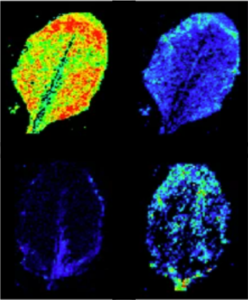
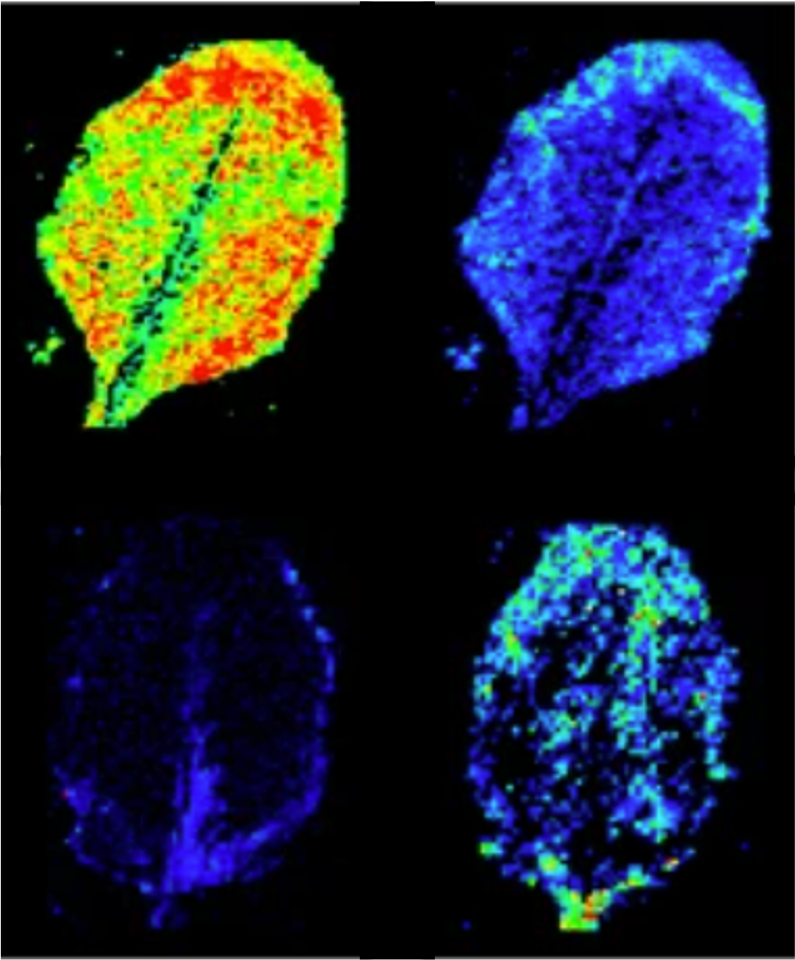
Metabolic footprint of epiphytic bacteria on Arabidopsis thaliana leaves | The ISME Journal